¶ OME-NGFF Converter by Glencoe
Glencoe has released a statement regarding the conversion of complex formats to pyramidal ome.tiff
https://www.glencoesoftware.com/blog/2019/12/09/converting-whole-slide-images-to-OME-TIFF.html
In 2022, Glencoe released a standalone application which makes it easy to convert any to the OME-NGFF file format, or to OME-TIFF:
Because QuPath do not read OME-NGFF dataset, we recommend to convert to OME-TIFF currently (2022/11/11). This can be done with the OME-NGFF Converter.
There are two issues however with the Glencoe conversion which makes other options still valid:
- If a single plane of an image is too big, and the downscaling factor is too big, the Glencoe conversion will likely fail. More details on the forum.
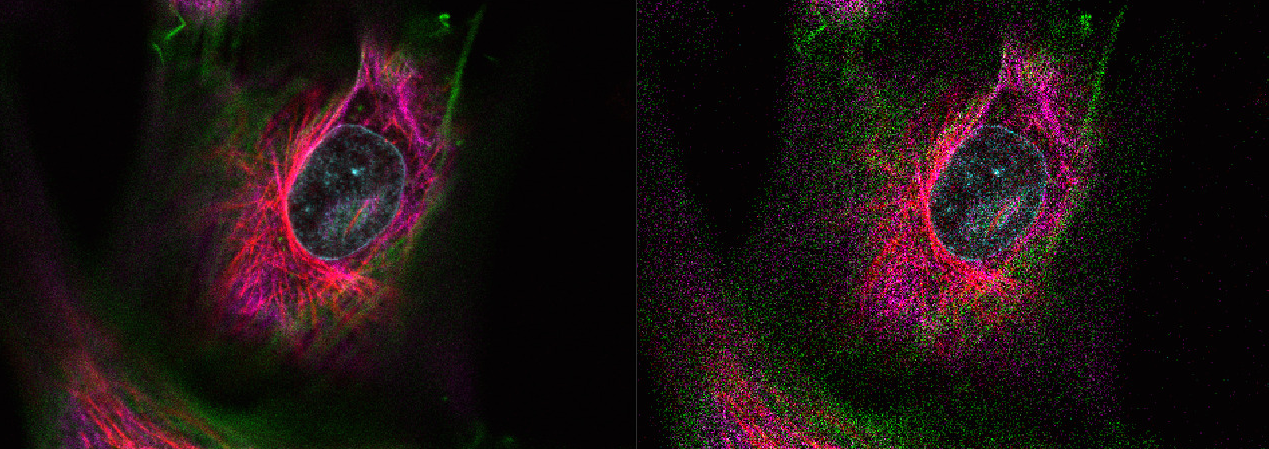
- The downscaling is using nearest neighbor interpolation instead of averaging over underlying pixels. This can lead to drastically degraded images with high downscaling factor and low SNR:
¶ Kheops
Kheops is a tool written by the BIOP which can be an alternative to the OME converter. It is highly efficient for exporting many images in parallel, and computes an averaging over pixels when downscaling. It is also very memory efficient in its process.
HOWEVER, currently all metadata except the pixel size are lost. Also, it won't convert MRXS files.
¶ Legacy workflows: Fiji scripts and manual install
Before the above tools (Glencoe, Kheops) were available, there was a way to convert any bio-formats supported file or MRXS to OME-TIFF, but it requires some extra installation. The advantage over other options is not clear.
¶ Workflow Summary
The Glencoe protocol describes a two-step process.
- Convert vendor data to 'raw' data using
bioformats2raw - Convert 'raw' data to ome.tiff using
raw2ometiff
The need for the two separate protocols seems to stem from licensing and dependencies on separate vendor-specific libraries.
Because this protocol makes use of very modern file storage and compression systems, there is a dependency on a tool called
c-blosc
¶ Installing c-blosc
¶ In windows
The procedure is described in https://github.com/glencoesoftware/bioformats2raw (and re-summarized here):
- download the latest pre-built blosc DLLs available from the Fiji project.
- Rename the downloaded DLL to
blosc.dlland place it in a fixed location (for instanceC:\path\to\blosc\folder) - then set a system environment variable named
JAVA_OPTSwhich value is set to-Djna.library.path=C:\path\to\blosc\folder
¶ Not windows
c-blosc can also be compiled, see legacy documentation below and readme of https://github.com/glencoesoftware/bioformats2raw
¶ Installing bioformats2raw
Unzip this somewhere
https://github.com/glencoesoftware/bioformats2raw/releases
¶ Installing raw2ometiff
Unzip this somewhere
https://github.com/glencoesoftware/raw2ometiff/releases
¶ Adding to the PATH
Add the two unzipped bin folders to your PATH system environment variable so you can call these two functions from anywhere. Alternatively, you can set the path explicitly in the provided groovy script:
//bf2rawPath = "bioformats2raw.bat"
//raw2ometiffPath = "raw2ometiff.bat"
bf2rawPath = "C:/whatever/.../bioformats2raw.bat"
raw2ometiffPath = "C:/whatever/.../raw2ometiff.bat"
¶ Using this workflow within Fiji
You can use the bf2ometiff gist and click batch to batch convert your files.
¶ Using This workflow with the command line
Example: Dataset called D:\ToConvert\Kidney.mrxs which is RGB
bioformats2raw.bat --resolutions=4 D:\ToConvert\Kidney.mrxs D:\ToConvert\Kidney
Other arguments are available with bioformats2raw.bat --help
Converting to OME.TIFF
raw2ometiff.bat --compression="JPEG-2000" --rgb D:\ToConvert\kidney D:\ToConvert\Kidney.ome.tiff
RGB images must be explictely concerted using the
--rgbflag. Otherwise it will be a 3 channel image.
¶ Installing c-blosc from source
¶ Dependencies
Download Build Tools for Visual Studio 2019
Install the packages
MSVC v142 - VS 2019 C++ x64/x86 build toolsWindows 10 SDK (10.0.18362.0)
Download CMake (msi installer) and install with checking Add CMAKE Path to all users.
If you followed the StarDist with gpu-tools installation, you have all the prerequisites except for CMake. Otherwise please follow the installation of the StarDist Prerequisites
¶ Installation
- Clone https://github.com/Blosc/c-blosc
- Create a folder in your
C:\drive calledDev Tools - Open a command line and do the following
cd c-blosc
mkdir build
cmake -DCMAKE_INSTALL_PREFIX="C:\Dev Tools"
cmake --build . --target install