OMERO.Tables cannot be created manually in the web interface. A script or .csv file is necessary to be able to create an OMERO.Table. You can add tables on an image, a dataset or a project.
¶ Create OMERO.Tables from a .csv file
¶ Description
OMERO.tables unifies the storage of columnar data from various sources, such as automated analysis results or script-based processing, and makes them dynamically available within OMERO.
¶ Ressources
Dataset: Copy the folder called
NEDDfrom:WIN: \\sv-nas1.rcp.epfl.ch\ptbiop-raw\public\1-Training\OMERO\demo dataset_OMERO-Fiji
MAC: smb://sv-nas1.rcp.epfl.ch/ptbiop-raw/public/1-Training/OMERO/demo dataset_OMERO-Fiji
CSV file: Copy the file
four-NEDD-images.csvfrom:WIN: \\sv-nas1.rcp.epfl.ch\ptbiop-raw\public\1-Training\OMERO\demo extra files
MAC: smb://sv-nas1.rcp.epfl.ch/ptbiop-raw/public/1-Training/OMERO/demo extra files
¶ Step-by-Step:
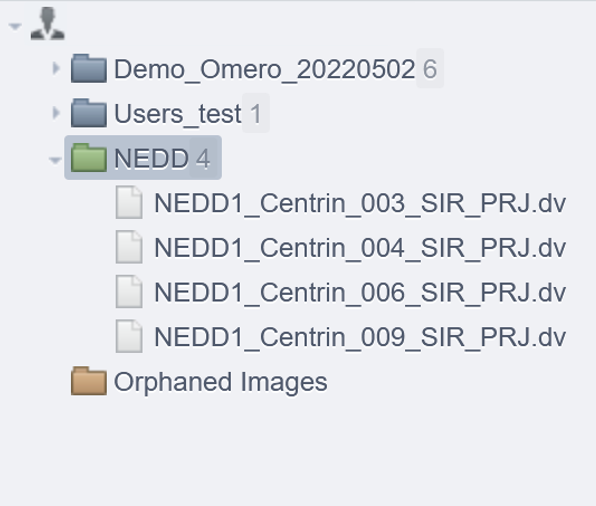
- Upload the folder called "NEDD" to OMERO as a Dataset. See Import images page to know how to import data on OMERO.
- In the webclient, and if it is not already the case, name this dataset as
NEDD.
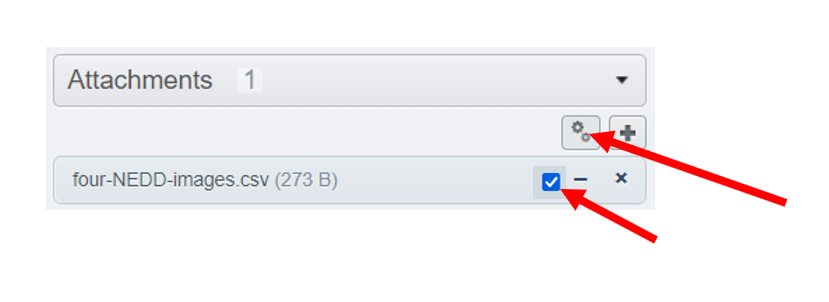
- Attach the
four-NEDD-images.csvfile to yourNEDDDataset as an attachment. See annotation page to know how add an attachment. - Select the
NEDDdataset - Select the previous csv file
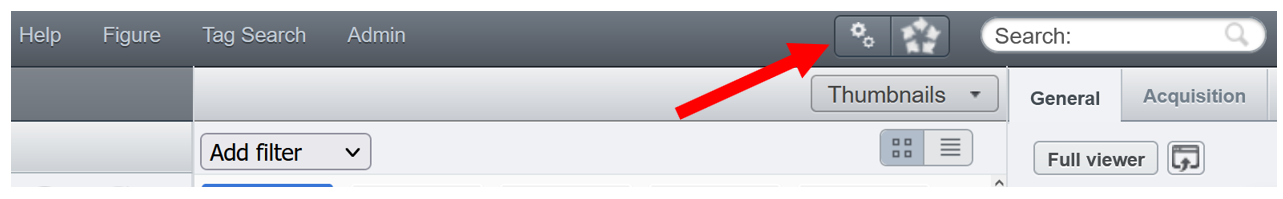
- Click on the gears button from the top menu bar.
- Click on
import_scripts → Populate Metadata...
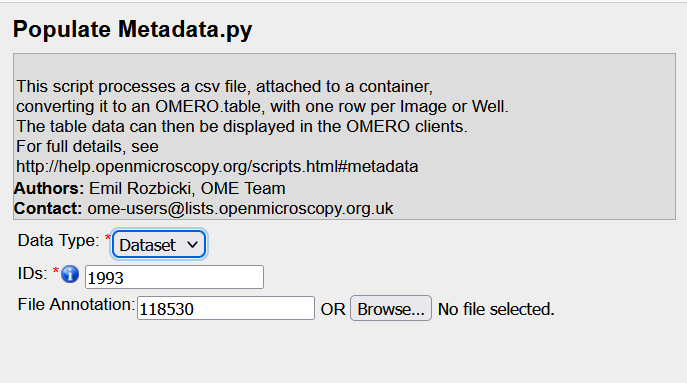
- Check that the
IDsfield shows the ID of the dataset holding the images to annotate. - Check that the
File Annotationstates the ID of the attachment (Annotation ID field when you pass your mouse over the corresponding attachment). In case you did not select the attachment in the previous step, you can enter the attachment ID here.

-
Click on
Run Script. -
Select the
NEDDdataset from the left-hand pane. Click “Attachments” harmonica in the right panel. -
A new file called
bulk_annotationsis attached to yourNEDDdataset. This is your newly created OMERO.table, that can be opened from the web by hitting the "eye" button. It contains in each line a dynamic link to the corresponding image in the webclient.
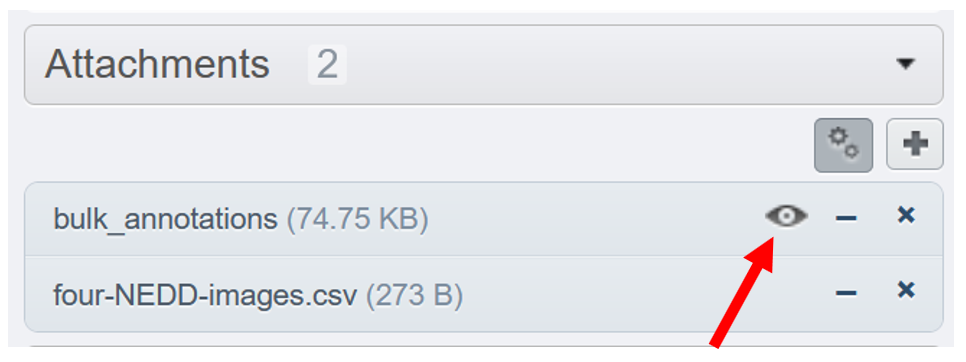
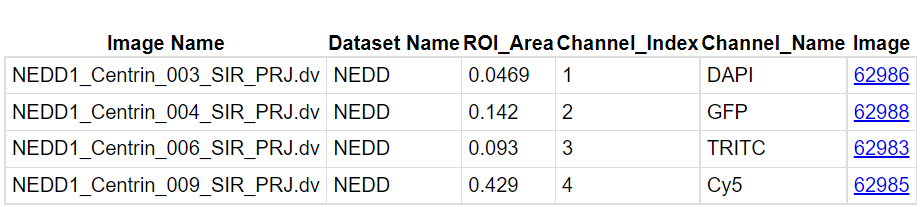
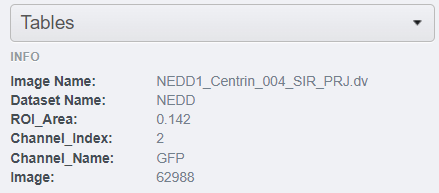
- Back to webclient's main page, clic on one of the images from the
NEDDdataset without opening it with the iviewer. Look at the "Tables" harmonica from the right-hand pane. Observe that all the statistics from the OMERO.table that apply to the selected image are gathered here.
¶ Having a look to the csv file
To be compatible with Populate_metadata.py script, the csv file has to be formatted as following.
- A first header row to format each column as string (s), double (d), long (l) and boolean (b)
- A second header row to give a name to each column (without any space in names EXCEPT for the two following headers).
- A column containing each image name (with header Image Name)
- (optional) A column containing the name of the dataset that images referred to (with header Dataset Name)
- Other columns with data
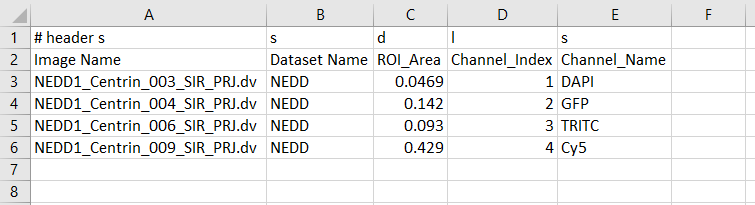
Do not forget the # header in the first cell !
More info on the official documentation .
Be careful : You will not be able to add fields in OMERO.parade coming this csv file because it is NOT compatible. If you want to use OMERO.parade with csv files, please read the OMERO.parade page.
¶ Create OMERO.Tables from a Fiji script
You can find some code templates on our github repo showing you how to deal with OMERO.tables
You can also follow the Segment and analyze OMERO dataset with scripts example if you want to have a full workflow.
¶ Tables on dataset level
Having OMERO.tables at the dataset level has some advantages if they are correctly formatted.
-
Summarize metrics for each image within the dataset in one row.
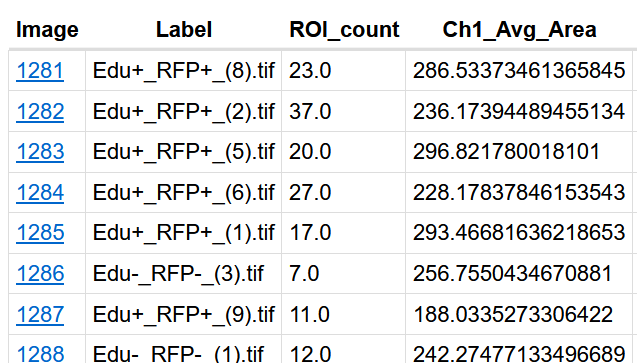
-
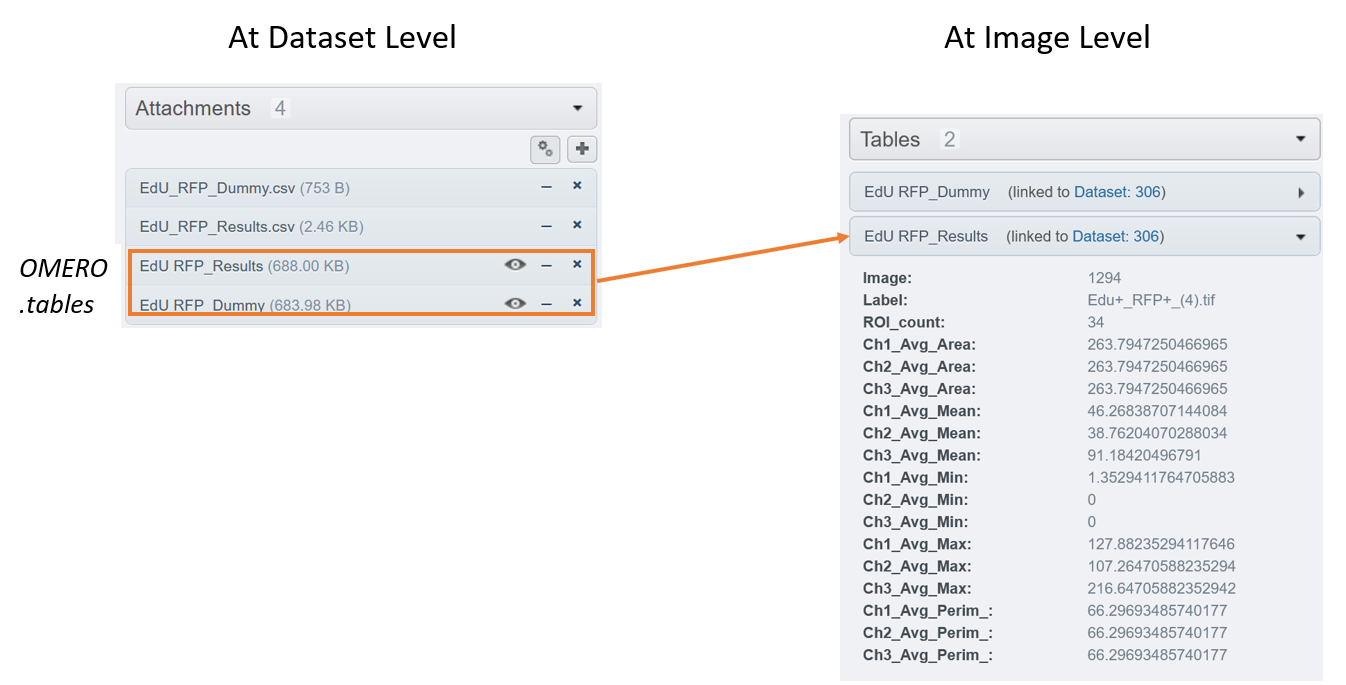
Display the corresponding summary at the image level, under the
Tableharmonica.
- Use this table in OMERO.parade for data mining.
To be usable, each table should have one row per image within the dataset. Please refer to the above chapters to know how to create an OMERO.table.
Having one or more OMERO.tables at the dataset level is fully compatible with OMERO.tables at the image level.
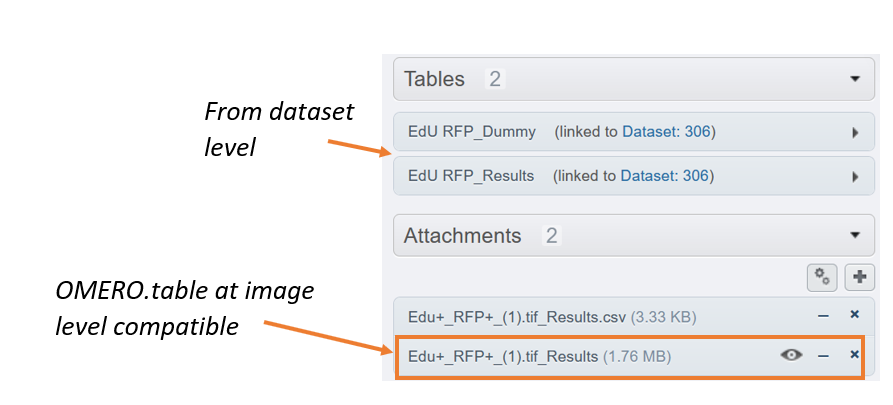
You can add more than one OMERO.table at the dataset level and each of them will be summarized at the image level. But you will not be able to use OMERO.parade anymore because it does not support multiple OMERO.tables.